Tryptophan (Trp) Operon – Structure, Regulatory, Mechanism, Function
The trp operon of E. coli controls the biosynthesis of tryptophan in the cell from the initial precursor chorismic acid. This operon contains genes for the production of five proteins which are used to produce three enzymes. The products of the E and D genes form a multimeric protein comprised of two copies of each protein to produce the enzyme anthranilate synthetase. This enzyme catalyzes the first two reactions in the tryptophan pathway. The next enzyme, which is responsible for catalyzing the next two steps in the pathway is indole glycerolphosphate synthetase and it is the product of the C locus. The final step in the reaction is the pathway produces tryptophan from indole-glycerol phosphate and serine. This single step is catalyzed by tryptophan synthetase, an enzyme that is a multimer of two proteins that are the product of the B and A genes.
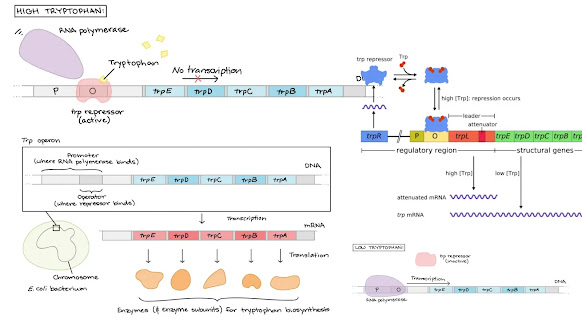
As with all operons, the trp operon consists of the
repressor, promoter, operator and the structural genes. In this system, though,
unlike the lac operon, the gene for the repressor is not adjacent to the
promoter, but rather is located in another part of the E. coli genome. Another
difference is that the operator resides entirely within the promoter
The trp operon in E. coli contains five genes that produce
proteins that are used in the production of the amino acid tryptophan when
needed by the cell. When tryptophan levels in the cell are high, tryptophan
binds to the trp operon repressor protein, which activates it. The activated
repressor then binds to the operator, preventing RNA polymerase from binding
and transcribing the operon. However, when tryptophan concentrations in the
cell are low, it doesn't bind to the repressor, preventing it from binding to
the operator, and allowing transcription until the terminator after the trpA
gene is reached. The trp operon is also regulated by the amount of useable trp
tRNA present. Upon start of transcription, the leader peptide, encoded by the
trpL gene, will begin to be transcribed. Because this peptide contains two trp
residues next to each other, and trp is a relatively uncommon amino acid, if
there is a low concentration of trp tRNA in the cell, it can cause the leader
peptide to stall during transcription. This allows for the section of mRNA
immediately after the stalled ribosome to form the anti-termination hairpin.
This hairpin prevents the formation of the terminal hairpin that contains a
termination sequence that would stop transcription after the leader peptide.
Because the anti-termination hairpin is allowed to form, transcription of the
rest of the operon can continue. However, when the cell contains a high
concentration of trp tRNA, the transcription does not stall, which allows for
the formation of the transcription terminator to form before the rest of the
genes in the operon, preveinting their transcription. The trpE and trpD genes
encode for anthranilate synthase components 1 and 2 respectively. These combine
to create anthranilate synthase, which produces anthranilate and pyruvate from
chorismate. The trpC gene encodes the tryptophan biosynthesis protein that
takes the anthranilate from the previous protein and converts it in two steps
to indole-3-glycerol. Finally, the trpB and trpA genes encode for tryptophan
beta and alpha subunits respectively. Two of each subunit come together to form
tryptophan synthase. This protein then takes the previous compound, as well as
a molecule of L-serine, and catalzes their conversion into tryptophan, as well
as water and D-glyceraldehyde-3-phosphate.
The tryptophan operon is responsible for the production of
the amino acid tryptophan, whose synthesis occurs in five steps, each requiring
a particular enzyme. In E. coli, these enzymes are translated from a single
polycistronic mRNA. Adjacent to the enzyme coding sequences in the DNA are a
promoter, an operator, and two regions called the leader and the attenuator.
The leader and attenuator sequences are transcribed. Another gene (trpR)
encoding a repressor is located some distance from this gene cluster.
Tryptophan (Trp) Operon Definition
The trp operon of E. coli controls the biosynthesis of
tryptophan in the cell from the initial precursor chorismic acid. This operon
contains genes for the production of five proteins which are used to produce
three enzymes. The products of the E and D genes form a multimeric protein
comprised of two copies of each protein to produce the enzyme anthranilate
synthetase. This enzyme catalyzes the first two reactions in the tryptophan
pathway. The next enzyme, which is responsible for catalyzing the next two
steps in the pathway is indole glycerolphosphate synthetase and it is the
product of the C locus. The final step in the reaction is the pathway produces
tryptophan from indole-glycerol phosphate and serine. This single step is
catalyzed by tryptophan synthetase, an enzyme that is a multimer of two
proteins that are the product of the B and A genes.
As with all operons, the trp operon consists of the repressor, promoter, operator and the structural genes. In this system, though, unlike the lac operon, the gene for the repressor is not adjacent to the promoter, but rather is located in another part of the E. coli genome. Another difference is that the operator resides entirely within the promoter
- A collection of genes that are transcribed together encode the components for tryptophan synthesis.
- The E. coli trp operon is a cluster of genes that code for biosynthetic enzymes for the amino acid tryptophan.
- When tryptophan levels are low, the trp operon is expressed (turned “on”) and when they are high, it is repressed (turned “off”).
- The trp repressor controls the trp operon. When coupled to tryptophan, the trp repressor inhibits operon expression.
- Attenuation also controls the tryptophan biosynthesis (a mechanism based on coupling of transcription and translation).
- Bacteria such as Escherichia coli (a beneficial gut flora) require amino acids to exist because, like humans, they must construct proteins. Tryptophan is one of the amino acids they require.
- If tryptophan is present in the environment, E. coli will utilise it to produce proteins. However, E. coli can also produce tryptophan by the use of enzymes expressed by five genes. These five genes are positioned next to one another in the trp operon.
- If tryptophan is present in the environment, E. coli bacteria do not need to generate it; hence, transcription of the trp operon genes is “turned off.” In contrast, when tryptophan is scarce, the operon is activated, the genes are transcribed, biosynthetic enzymes are generated, and more tryptophan is created.
- This operon exemplifies repressible negative gene expression regulation.
- In the presence of tryptophan, the repressor protein attaches to the operator (repressing transcription) and is released from the operon in the absence of tryptophan (allowing transcription to proceed).
- The trp operon additionally uses attenuation, a second negative feedback control mechanism, to regulate the expression of the operon.
- The trp operon contains five structural genes that encode the enzymes required to produce tryptophan: trpE, trpD, trpC, trpB, and trpA.
- Additionally, it contains the repressive regulator gene trpR. When tryptophan is present, the trpR protein binds to the operator, preventing RNA polymerase from transcribing the trp operon.
This operon possesses two levels of regulation:
1. Repression: works at the transcription initiation level
2. Attenuation: works at the transcription termination level
Structure of the trp operon
The trp operon consists of five genes encoding enzymes required for tryptophan production, as well as a promoter (RNA polymerase binding site) and an operator (binding site for a repressor protein). The trp operon’s genes are transcribed as a single mRNA.
Structural genes
Five structural genes comprise Trp operon. Their functions
are:
- TrpE: Encodes the Anthranilate synthase enzyme I. Anthranilate is produced by anthranilate synthase.
- TrpD: Anthranilate synthase II is encoded by TrpD. Compatible with TrpE.
- TrpC: TrpC is responsible for encoding the enzymes N-5′-Phosphoribosyl anthranilate isomerase and Indole-3-glycerolphosphate synthase. First, the domain of phosphoribosylanthranilate isomerase converts N-(5-phospho—D-ribosyl)anthranilate into 1-(2-carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate. On the same protein, the Indole-3-glycerol-phosphate synthase converts the product into (1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate.
- trpB: trpB encodes the component of the enzyme tryptophan synthase-B.
- trpA: trpA encodes the component of the enzyme tryptophan synthase-A.
|
trp Operon Gene |
Gene Function |
|
P/O |
Promoter; operator sequence is
found in the promoter |
|
trp L |
Leader sequence; attenuator (A)
sequence is found in the leader |
|
trp E |
Gene for anthranilate
synthetase subunit |
|
trp D |
Gene for anthranilate
synthetase subunit |
|
trp C |
Gene for glycerolphosphate
synthetase |
|
trp B |
Gene for tryptophan synthetase
subunit |
|
trp A |
Gene for tryptophan synthetase
subunit |
Regulatory DNA sequences
Promoter Gene
- It refers to the region of a bacterial chromosome containing a particular nucleotide sequence to which an RNA polymerase can bind to commence transcription.
- The association of a repressor protein with the operator blocks the association of RNA polymerase with the promoter region, hence stopping transcription.
Operator Gene
- It is the precise sequence of nucleotides on the chromosomal DNA of E. coli to which a repressor protein can bind in the presence of an effector molecule or tryptophan.
- The tryptophan molecule functions as a corepressor to aid in the activation of the aporepressor protein.
Regulatory Gene
- A regulatory gene on the chromosome encodes the trp repressor protein that recognises the operator sequence in the tryptophan operon.
- The repressor protein activates the operon system when the surrounding trp concentration is low and deactivates the system when the trp concentration is high.
- Consequently, a repressor protein is related with the synthesis of five gene products, which is dependent on the surrounding quantity of tryptophan.
- Repressor activity is dependent on the presence or absence of a corepressor or effector molecule (tryptophan).
Attenuator Region
- It is located between the operator region and the structural genes.
- The attenuator region consists of 160-bp leader sequences that regulate transcription via attenuation.
- Its mechanism consists of generating dimers within the bacterial cell at sufficient levels of tryptophan in order to reduce transcriptional effectiveness.
- In contrast, the RNAp can cross the attenuator region and transcribe the genes required for trp synthesis at low tryptophan levels.
Structure of the trp operon
- Turning the operon “on” and “off”
- The trp repressor is a regulatory protein that recognises the operator. When the repressor binds to the operator’s DNA, it prevents the operon from being transcribed by physically interfering with the transcription enzyme, RNA polymerase.
- The trp repressor protein is encoded by the trpR gene. This gene is located elsewhere on the bacterial chromosome, with its own promoter and other regulatory regions, and is not part of the trp operon.
- Not always does the trp repressor attach to DNA. Instead, it exclusively binds and inhibits transcription in the presence of tryptophan.
- When tryptophan is present, it binds to repressor molecules and alters their structure, activating them.
- A corepressor is a tiny chemical, such as trytophan, that flips a repressor into its active state.
- However, when there is minimal tryptophan in the cell, the trp repressor is inactive (because no tryptophan is available to bind to and activate it).
- It does not bind to DNA or inhibit transcription, allowing RNA polymerase to transcribe the trp operon.
- The trp repressor in this system serves as both a sensor and a switch. If tryptophan is already present in large concentrations, it switches the operon to the “off” state, preventing the production of superfluous biosynthetic enzymes.
Repressor or derepression mechanism
Attenuation mechanism
- Repression of Trp Operon
- The repressive regulation is detrimental.
- The trpR gene, which codes for the repressor of the trp operon, is positioned far from the operon itself.
- P1 refers to the major promoter region, which includes the operator (O) region.
- P2 is a weak promoter found at the farthest end of the trpD gene from the operator. Its role is to elevate the basal transcription levels of trpC, trpB, and trpA.
- Two termination areas, t and t’, are located downstream of the trpA gene. The trpL indicates a region containing an mRNA leader sequence (162 nucleotides in length).
- It functions as a co-repressor. RNA polymerase attaching to the promoter stimulates transcription of structural genes in its absence.
- When tryptophan is present, it forms a corepressor/repressor complex with the repressor.
- This complex binds to the operator sequence and inhibits the initiation of transcription by RNA polymerase.
- In the presence of tryptophan (repressed state), RNA polymerase activity is approximately 70 times slower than in the absence of tryptophan (de-repressed state).
- In trpR mutants, there is no functional repressor, yet the addition of tryptophan can still cause a 10 fold reduction in transcription rate due to attenuation.
Mechanism
- It occurs when the concentration of trp in the surrounding medium is high.
- In this instance, the TrpR gene of the tryptophan operon produces apo-repressor (inactive) protein, which cannot bind to the operator region on its own.
- The apo-repressor protein activates and inhibits the RNA polymerase in the presence of a corepressor or tryptophan.
- Thus, RNAP cannot activate the transcription of structural genes and inhibits enzymes required for trp synthesis. At a high tryptophan concentration, the relevant RNA polymerase cannot bind to the operator gene or transcribe structural genes.
- Expression of the trp operon when tryptophan is available suggests that the operon system will end transcription.
- The repression of the trp operon is therefore mediated by a complex composed of an allosteric repressor and an effector molecule. Tryptophan acts as a corepressor or effector molecule that converts the inactive apo-repressor into an active repressor, which binds to the operator sequence to inhibit gene expression.
Derepression of Trp Operon
- It occurs when there are low quantities of tryptophan in the environment.
- Due to a deficiency in effector molecules, the active repressor protein will detach from the operator region.
- After separation, the repressor is rendered inactive and without function. Consequently, RNA polymerase is liberated to continue transcribing structural genes to create tryptophan.
- Expression of the trp operon in the absence of tryptophan results in the activation of the operon system to undertake transcription of structural genes by RNA polymerase.
- Thus, derepression is brought about by the dissociation of repressor protein due to the absence of trp or effector molecules, which can form an active complex.
Attenuation of trp Operon
- It is the second part of the trp operon that controls how the gene works. The trpL gene or attenuator controls this part.
- The expression of a gene is controlled by an attenuation mechanism that is part of a leader sequence.
- It is made up of a polypeptide sequence and an attenuator (contains palindromic sequences).
- Once the bacterial DNA is turned into mRNA, the palindromic sequences in the attenuator sequence can pair up and form dimers.
- In the leader sequence, there are four domains. Domain 3 can pair with either domain 2 or 4, and domain 1 can pair with domain 2. Also, there are two trp residues in it.
- When domains 2 and 3 work together, this is called antitermination. On the other hand, when domains 3 and 4 come together, trp biosynthesis stops.
- Domain-4, which is also called an attenuator, is needed to stop transcription because it is the only thing that can help form a stem-loop.
- The way the attenuation works depends on how the ribosomes pair up and how much tryptophan is in a bacterial cell.
At low tryptophan level
- Domain 1 of the mRNA transcript is where the ribosome sits.
- Then, because there isn’t much tryptophan, it translates the mRNA very slowly. Because the ribosome stops at domain-1, domain-3 interacts with domain-2.
- In this case, there won’t be a stem and loop structure. So, the transcription may continue to make enzymes that are needed for making trp.
At high tryptophan level
- When there is a lot of tryptophan in the cell, the ribosome moves quickly through domain 1 and stops at domain 2.
- Because of this, domain-3 connects to domain-4 and helps make a hair-loop structure.
- The dimerization of domains 3 and 4 makes the RNAp fall off and stops mRNA from transcribing genes that make enzymes for trp biosynthesis.
- Due to the pairing of self-complementary sequences, an attenuator acts as a barrier when there is a lot of trp.
The Arabinose Operon: An Example Of Positive And Negative control
- The arabinose operon is hard to understand because it is so complicated. Only the basic information is given below.
- Arabinose is broken down (catabolized) by the structural genes araB, araA, and araD.
- Arabinose structural gene transcription can be both repressed (negative control) and stimulated (positive control) by the regulator protein araC.
- In the absence of arabinose, the araC protein binds to araO2 (an operator) and araI (an insertion site). It is believed that this leads to a change in the secondary structure of PBAD (the promoter for structural genes) that prohibits or interferes with RNA polymerase binding at PBAD.
- In the presence of arabinose and cyclic AMP, araC becomes a transcriptional activator. In this instance, an arabinose-araC protein complex and a cyclic AMP-CAP protein complex with binding sites at the araI site are involved. The “model” proposes that the two complexes influence the binding of RNA polymerase to PBAD in a beneficial manner.
- The Pathway and Enzymes of Tryptophan Biosynthesis
- The biosynthetic process for tryptophan begins with chorismate, the common precursor for the three aromatic amino acids.
- Chorismate is also a precursor to a number of minor aromatic metabolites, such as p-aminobenzoic acid, a component of folic acid.
- The molecular pathways from chorismate to tryptophan, as well as the seven polypeptide domains that catalyse these pathways.
- A tetrameric enzyme complex comprised of two TrpE and two TrpG–TrpD polypeptides catalyses the synthesis of anthranilate from chorismate and phosphoribosyl anthranilate from anthranilate.
- Even though L-glutamine is the amino group donor of choice during the synthesis of anthranilate (o-aminobenzoate) from chorismate, ammonia can be used by the complex or the TrpE polypeptide alone as an alternative source of this amino group.
- Glutamine consumption requires the TrpG glutamine amidotransferase domain.
- The TrpD domain provides 5-phosphoribosyl-1-pyrophosphate (PRPP) to the side chain of phosphoribosyl anthranilate during the conversion of anthranilate to phosphoribosyl anthranilate.
- Anthranilate phosphoribosyltransferase, the TrpF domain, then rearranges phosphoribosyl anthranilate to create 1-(o-carboxyphenylamino)-1-deoxyribulose-5-phosphate (CdRP).
- The carboxyl group of CdRP is subsequently removed, and the pyrrole ring of the indole moiety is produced, resulting in indole-3-glycerol phosphate, the next stage in the route.
- The TrpC domain of indoleglycerol phosphate synthase catalyses the latter process.
- The TrpA polypeptide of the tryptophan synthase enzyme complex converts indole glycerol phosphate to indole. This tetrameric complex consists of two molecules of TrpA and two molecules of TrpB.
- L-tryptophan is formed by condensing indole with a pyridoxal phosphate derivative of L-serine; the final reaction is catalysed by the TrpB polypeptide of the tryptophan synthase complex.
- The synthesis of tryptophan from chorismate requires the chemicals L-glutamine, phosphoribosyl-1-pyrophosphate, L-serine, and pyridoxal phosphate, which are the end products of four more metabolic processes.
- The amino group of anthranilate comes from glutamine. Two carbon atoms of the pyrrole ring of indole come from phosporibosyl pyrophosphate. The alanyl side chain of tryptophan comes from L-serine. Pyridoxal phosphate is the coenzyme that is needed to activate L-serine and speed up the final reaction in the formation of tryptophan.
Structure/Function of Tryptophan Biosynthetic Enzymes
- The mechanism of enzymatic catalysis of each of the tryptophan biosynthetic processes has been studied, as well as the active site residues of each biosynthetic protein and protein domain.
- The three-dimensional structure of the Salmonella typhimurium tryptophan synthase enzyme complex and the structures of complexes harbouring mutant protein variations have been determined.
- E. coli’s bifunctional phosphoribosyl anthranilate isomerase-indoleglycerol phosphate synthase has also been characterised in three dimensions.
- These structures have indicated that the polypeptide domains of TrpA, TrpC, and TrpF have similar structures of the / triosephosphate isomerase (TIM) barrel type, suggesting the idea that they developed from one another or a shared ancestor.
- The active site of the TrpA polypeptide and the active site of the TrpB polypeptide are connected by a tunnel, according to structural studies of the tryptophan synthase enzyme complex.
- Indole is transported down this tunnel from the TrpA active site to the TrpB active site, where it is condensed with serine.
- Studies of this enzyme complex have also revealed characteristics of the complex that explain the mutual activation of each polypeptide upon creation of a complex with a heterologous polypeptide.
Trp Operon Summary
- In repressible operons, such as the tryptophan (or trp) operon, the repressor cannot bind to the operator DNA alone.
- In contrast, the repressor is only active as a DNA-binding protein when complexed with a particular component, such as tryptophan, which serves as a corepressor.
- In the absence of tryptophan, the operator site is accessible to RNA polymerase, which then binds and transcribes the structural genes of the trp operon, resulting in the creation of the enzymes that manufacture tryptophan.
- The enzymes of the tryptophan synthesis pathway are no longer necessary once tryptophan becomes accessible.
- Under these conditions, the increased tryptophan concentration results in the development of the transcription-blocking tryptophan–repressor complex.
References
David Hames and Nigel Hooper (2005). Biochemistry. Third ed.
Taylor & Francis Group: New York.
Parija S.C. (2012). Textbook of Microbiology &
Immunology.(2 ed.). India: Elsevier India.
Lehninger Principles of Biochemistry: International Edition
by David L. Nelson (Author), Michael Cox
(Author)
Molecular Biology by Robert F. Weaver (Author)
Yanofsky, C. (2013). Tryptophan Operon of Escherichia coli.
Brenner’s Encyclopedia of Genetics, 221–223.
doi:10.1016/b978-0-12-374984-0.01676-4










.jpg)







No comments