Pulsed Field Gel Electrophoresis (PFGE) – Principle, Method, Applications
Pulsed-field gel electrophoresis (PFGE) is considered the
"gold standard" for bacteria typing. The method involves enzyme
restriction of bacteria DNA, separation of the restricted DNA bands using a
pulsed-field electrophoresis chamber, followed by clonal assignment of bacteria
based on PFGE banding patterns. Various PFGE protocols have been developed for
typing different bacteria, leading it to be one of the most widely used methods
for phylogenetic studies, food safety surveillance, infection control and
outbreak investigations. On the other hand, as PFGE is lengthy and labourious,
several PCR-based typing methods can be used as alternatives for research
purposes. Recently, matrix-assisted laser desorption/ionization time of flight
mass spectrometry (MALDI-TOF MS) and whole genome sequencing (WGS) have also
been proposed for bacteria typing. In fact, as WGS provides more information,
such as antimicrobial resistance and virulence of the tested bacteria in
comparison to PFGE, more and more laboratories are currently transitioning from
PFGE to WGS for bacteria typing. Nevertheless, PFGE will remain an affordable
and relevant technique for small laboratories and hospitals in years to come.
Pulsed Field Gel Electrophoresis (PFGE) is a technique used for the separation of large deoxyribonucleic acid (DNA) molecules by applying to a gel matrix an electric field that periodically changes direction. As DNA larger than 15-20kb migrating through a gel essentially moves together in a size-independent manner, the standard gel electrophoresis technique was unable to separate very large molecules of DNA effectively which led to the practice of pulsed field gel electrophoresis. In 1982, Schwartz introduced the concept that DNA molecules larger than 50 kb can be separated by using two alternating electric fields
Genotyping of microorganisms is very important in evaluating
the global evolution of the pathogens and studying their genetic relatedness to
determine their point source during epidemiological investigations. A variety
of genotyping methods exists for Staphylococcus aureus. Each has strengths and
weaknesses. These methods include pulse field gel electrophoresis (PFGE),
surface protein A typing (spa-typing), multi locus sequence typing (MLST), SCC
mec typing, plasmid profile analysis, restriction fragment length polymorphism
(RFLP), RFLP-Southern blot, Rep-PCR typing, Multilocus VNTR (Variable Number
Tandem Repeat) analysis (MLVA), and whole-genome DNA sequence typing.
S. aureus is one of the most important causes of
life-threatening bacterial infections in the industrialized world causing
infections both in the hospitals and the community. S. aureus is the second
most common overall cause of healthcare-associated infections reported to the
National Healthcare Safety Network, the most common cause of surgical site
infections, the leading cause of infections involving heart valves and cardiac
devices, and a leading cause of bacteremia and endocarditis. Additionally, S.
aureus routinely becomes resistant to many of the currently available
antibiotic therapies. Recently, the Centers for Disease Control and Prevention
estimated that 80,461 invasive MRSA infections and 11,285 related deaths
occurred in 2011 in the USA, even more than HIV/AIDS. Thus, a reproducible and
highly discriminatory genotyping technique to rapidly differentiate and type
these isolates is needed to prevent the illness and costs associated with these
infections. In addition, the availability of a sensitive genotyping method for
staphylococcal isolates is essential in understanding the epidemiological
evolution and outbreak of several antibiotic resistant strains including MRSA.
Among the various DNA-based methods available for genotyping
S. aureus and other bacterial pathogens, PFGE is often considered as a gold
standard due to its discriminatory power, reproducibility, and ease of
execution, data interpretation, cost, and availability. PFGE is a powerful
genotyping technique used for the separation of large DNA molecules (entire
genomic DNA) after digesting with unique restriction enzyme. First developed by
Schwartz et al. in yeast, PFGE is reported to be very sensitive, highly
reproducible with a very good discriminatory power in genotyping of S. aureus
isolates. PFGE involves the isolation of the intact chromosomal DNA by lysing
bacterial cells embedded in an agarose plug to avoid the mechanical shearing of
DNA molecules during the extraction. This is followed by digestion of the chromosomal
DNA within the agarose plug by a rare cutting restriction enzyme to produce ≥12
high-molecular weight DNA fragments. Finally, the digested DNA samples (10–800
kb) are subjected to separation by alternating the electric field between
spatially distinct pairs of electrodes. This will facilitate megabase (mb) size
DNAs to reorient and migrate at different speeds through the gel pores towards
the anode in a size dependent manner. The time required for reorientation is
also inversely proportional to the size of DNA fragment. Overall, this process
will achieve a good resolution of large DNA fragments in the agarose gel. The
obtained gel images will then be normalized and patterns of the DNA fragments
will be analyzed by BioNumerics Software following the criteria to interpret
PFGE patterns developed by Tenover et al. These patterns serve a virtual
barcode, which “types” the strains and allows for the determination whether
isolates are closely related.
Principle of Pulsed Field Gel Electrophoresis (PFGE)
- While in general small fragments can find their way through the gel matrix more easily than large DNA fragments, a threshold length exists above 30–50 kb where all large fragments will run at the same rate, and appear in a gel as a single large diffuse band.
- However, with periodic changing of field direction, the various lengths of DNA react to the change at differing rates. That is, larger pieces of DNA will be slower to realign their charge when field direction is changed, while smaller pieces will be quicker. Over the course of time with the consistent changing of directions, each band will begin to separate more and more even at very large lengths. Thus separation of very large DNA pieces using PFGE is made possible.
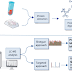
The Method of Pulsed Field Gel Electrophoresis (PFGE)
- The procedure for this technique is relatively similar to performing a standard gel electrophoresis except that instead of constantly running the voltage in one direction, the voltage is periodically switched among three directions; one that runs through the central axis of the gel and two that run at an angle of 60 degrees either side.
- The pulse times are equal for each direction resulting in a net forward migration of the DNA.
The major steps involved in Pulsed-field gel electrophoresis are:
- Lysis: First, the bacterial suspension is loaded into an agarose suspension. This is done to protect the chromosomal DNA from mechanical damage by immobilizing it into agarose blocks. Then the bacterial cells are lysed to release the DNA. The agarose-DNA suspension is also known as plug mold.
- Digestion of DNA: The bacterial DNA is treated with unusual cutting restriction enzymes so that it yields less number of larger size DNA fragments (in contrast to frequently used restriction enzymes used in RFLP which produces large number of smaller fragments).
- Electrophoresis: The larger pieces of DNA are subjected to pulse field gel electrophoresis by applying electric current and altering its direction at regular intervals (in contrast to the conventional agarose gel electrophoresis done to separate the smaller fragments where the current is applied in a single direction).
- Analysis: The fragments of different organisms generated by PFGE are compared to standards manually or by computer software like BioNumerics.
Applications of Pulsed Field Gel Electrophoresis (PFGE)
- Since, field gel electrophoresis allows the separation of DNA fragments containing up to 100,000 bp (100 kilobase pairs, or kbp), characterization of such large fragments has allowed construction of a physical map for the chromosomes from several bacterial species.
- PFGE may be used for genotyping or genetic fingerprinting.
- It is commonly considered a gold standard in epidemiological studies of pathogenic organisms.
- Subtyping has made it easier to discriminate among strains of Listeria monocytogenes and thus to link environmental or food isolates with clinical infections.
Advantages of Pulsed Field Gel Electrophoresis (PFGE)
- PFGE separates DNAs from a few kb to over 10 Mb pairs.
- PFGE subtyping has been successfully applied to the subtyping of many pathogenic bacteria and has high concordance with epidemiological relatedness.
- PFGE has been repeatedly shown to be more discriminating than methods such as ribotyping or multi- locus sequence typing for many bacteria.
- PFGE in the same basic format can be applied as a universal generic method for subtyping of bacteria. (Only the choice of the restriction enzyme and conditions for electrophoresis need to be optimized for each species.)
- DNA restriction patterns generated by PFGE are stable and reproducible.
Limitations of Pulsed Field Gel Electrophoresis (PFGE)
- Time-consuming.
- Requires trained and skilled technicians.
- Does not discriminate between all unrelated isolates.
- Pattern results vary slightly between technicians.
- Can’t optimize separation in every part of the gel at the same time.
- Don’t really know if bands of the same size are the same pieces of DNA.
- Bands are not independent.
- Change in one restriction site can mean more than one band change.
- “Relatedness” should be used as a guide, not true phylogenetic measure.
- Some strains cannot be typed by PFGE.
References
Bailey, W. R., Scott, E. G., Finegold, S. M., & Baron,
E. J. (1986). Bailey and Scott’s Diagnostic microbiology. St. Louis: Mosby.
Sastry A.S. & Bhat S.K. (2016). Essentials of Medical
Microbiology. New Delhi : Jaypee Brothers Medical Publishers.
Parija S.C. (2012). Textbook of Microbiology &
Immunology.(2 ed.). India: Elsevier India.

.jpg)









No comments